Optimize beta of the Gaussian kernel function by cross-validation
Source:R/nmfkc.R
nmfkc.kernel.beta.cv.Rdnmfkc.kernel.beta.cv selects the optimal beta parameter of the kernel function by applying cross-validation over a set of candidate values.
nmfkc.kernel.beta.cv(Y, Q = 2, U, V = NULL, beta = NULL, plot = TRUE, ...)Arguments
- Y
Observation matrix \(Y(P,N)\).
- Q
Rank of the basis matrix. Must satisfy \(Q \le \min(P,N)\).
- U
Covariate matrix \(U(K,N) = (u_1, \dots, u_N)\). Each row may be normalized in advance.
- V
Covariate matrix \(V(K,M) = (v_1, \dots, v_M)\), typically used for prediction. If
NULL, the default isU.- beta
A numeric vector of candidate kernel parameters to evaluate via cross-validation.
- plot
Logical. If TRUE (default), plots the objective function values for each candidate
beta.- ...
Additional arguments passed to
nmfkc.cv.
Value
A list with components:
- beta
The beta value that minimizes the cross-validation objective function.
- objfunc
Objective function values for each candidate
beta.
Examples
# install.packages("remotes")
# remotes::install_github("ksatohds/nmfkc")
# Example.
Y <- matrix(cars$dist,nrow=1)
U <- matrix(c(5,10,15,20,25),nrow=1)
V <- matrix(cars$speed,nrow=1)
nmfkc.kernel.beta.cv(Y,Q=1,U,V,beta=25:30/1000)
#> beta=0.025...
#> 0sec
#> beta=0.026...
#> 0sec
#> beta=0.027...
#> 0sec
#> beta=0.028...
#> 0sec
#> beta=0.029...
#> 0sec
#> beta=0.03...
#> 0sec
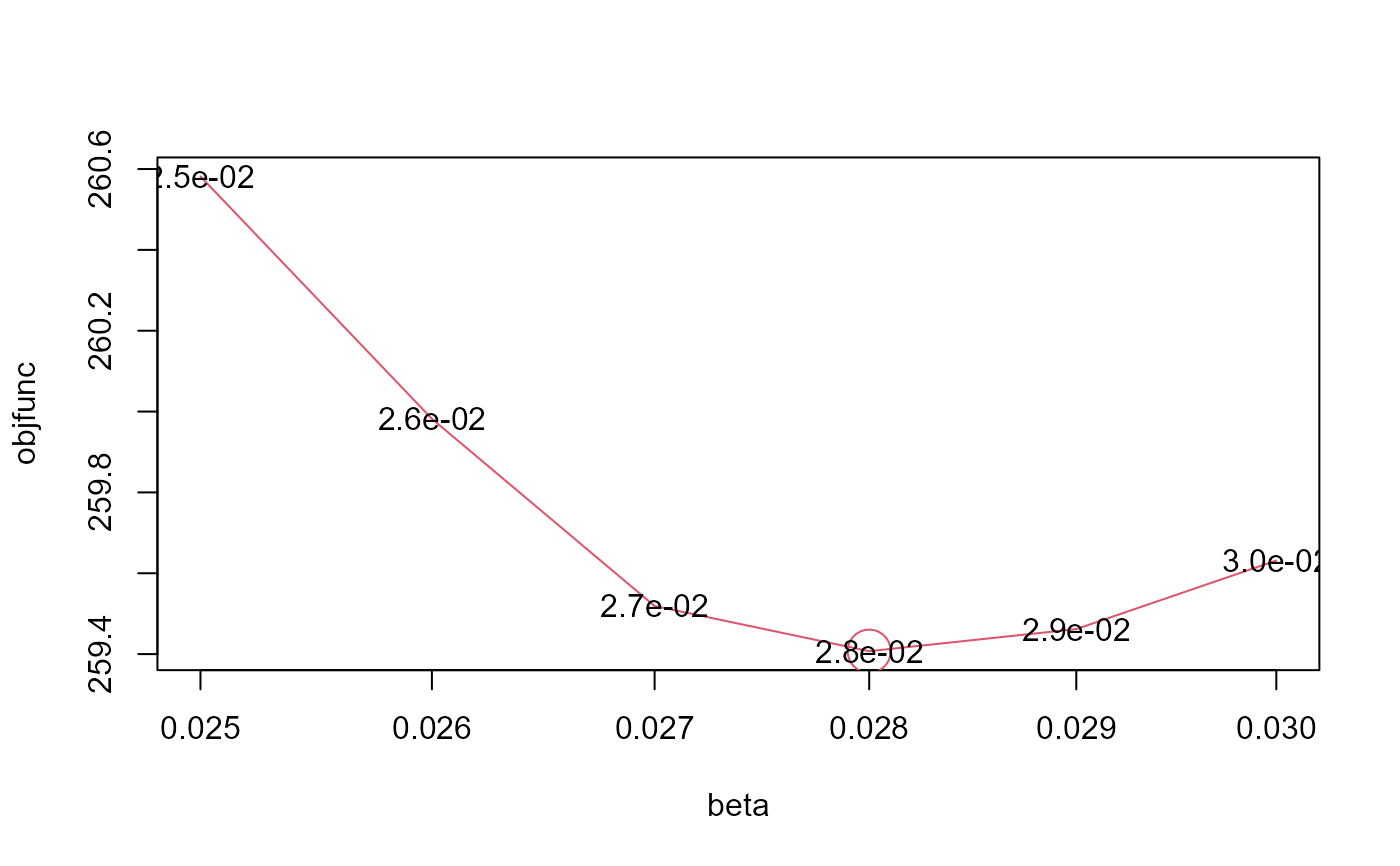 #> $beta
#> [1] 0.028
#>
#> $objfunc
#> 0.025 0.026 0.027 0.028 0.029 0.03
#> 260.5817 259.9821 259.5190 259.4072 259.4619 259.6314
#>
A <- nmfkc.kernel(U,V,beta=28/1000)
result <- nmfkc(Y,A,Q=1)
#> Y(1,50)~X(1,1)C(1,5)A(5,50)=XB(1,50)...
#> 0sec
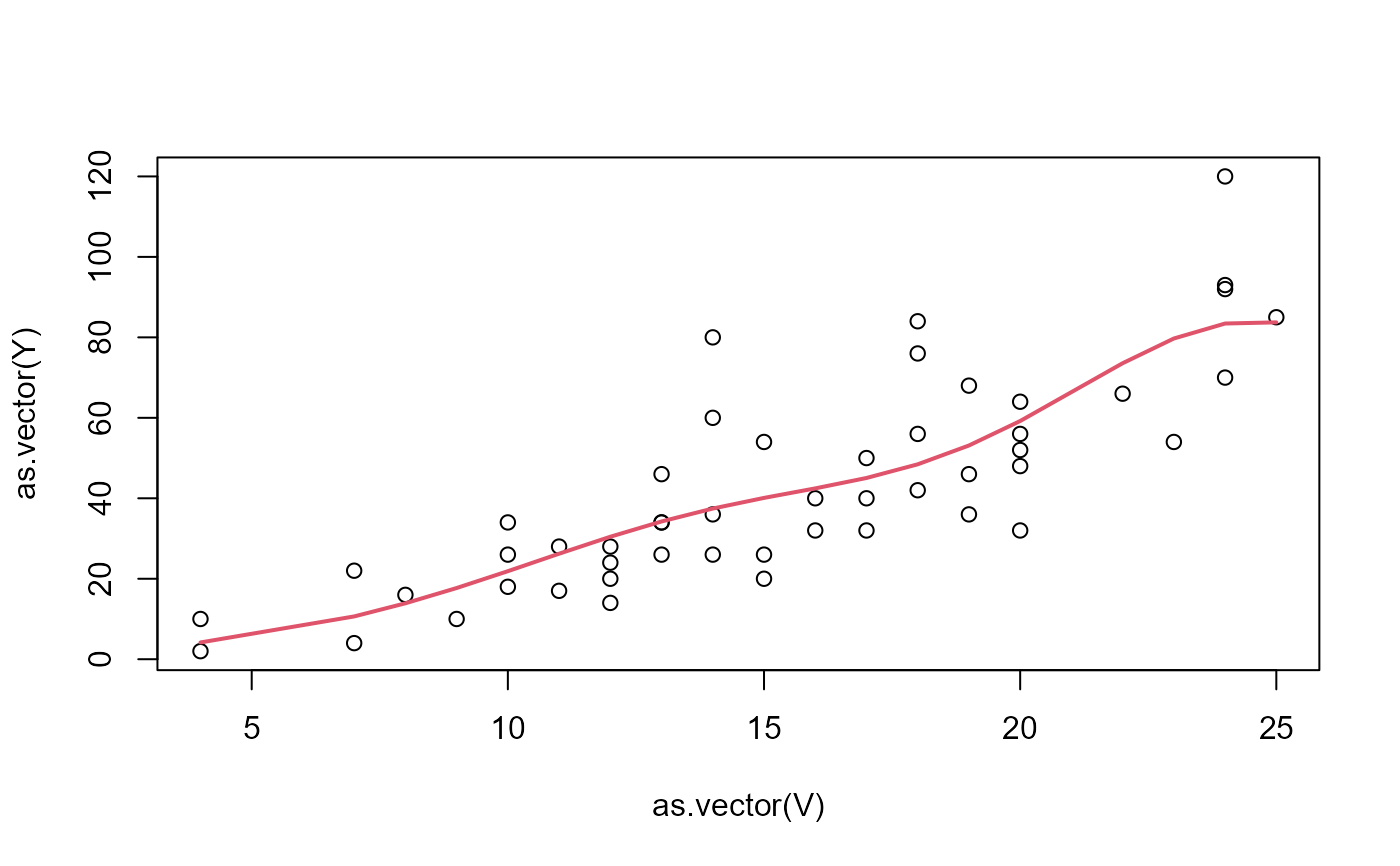
plot(as.vector(V),as.vector(Y))
lines(as.vector(V),as.vector(result$XB),col=2,lwd=2)
#> $beta
#> [1] 0.028
#>
#> $objfunc
#> 0.025 0.026 0.027 0.028 0.029 0.03
#> 260.5817 259.9821 259.5190 259.4072 259.4619 259.6314
#>
A <- nmfkc.kernel(U,V,beta=28/1000)
result <- nmfkc(Y,A,Q=1)
#> Y(1,50)~X(1,1)C(1,5)A(5,50)=XB(1,50)...
#> 0sec
plot(as.vector(V),as.vector(Y))
lines(as.vector(V),as.vector(result$XB),col=2,lwd=2)